Future Directions
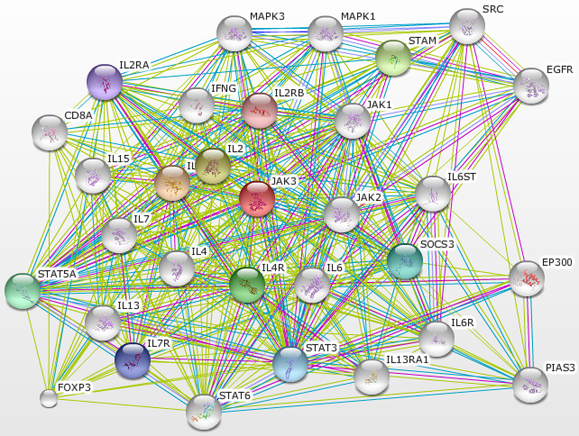
This page contains suggestions for experiments which could be performed to better understand JAK3 and the associated proteins. Figure 1 is an protein-protein interaction network produced using SMART. The interaction network centers on JAK3 and was taken out two degrees where we find the FOXP3 protein (seen in the lower left hand corner of the image).
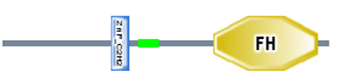
The FOXP3 (forkhead box P3) gene is located on the X chromosome on band p11.23 [2]. The FOXP3 protein contains a zinc finger domain and a forkhead domain (the blue rectangle and the gold colored "FH" polygon in figure 2 respectively). This protein is a transcriptional regulator that is necessary for the development and function of regulatory T-cells, which in turn are important for maintaining T-cell homeostasis [3]. When mutated FOXP3 can cause IPEX syndrome (immune disregulation, polyendocrinopathy, enteropathy x-linked syndrome), an autoimmune disease which is characterized by type I diabetes, thyroiditis, severe allergies, fatal infections, diarrhea, and dermatitis [3][4]. IPEX is x-linked recessive so males who inherit a mutated FOXP3 gene from their mother will have IPEX, while females need to inherit a copy from both parents in order to be affected [4]. Essentially this makes IPEX and SCID opposite health concerns, where IPEX presents with too many T-cells and SCID with too few or none at all.
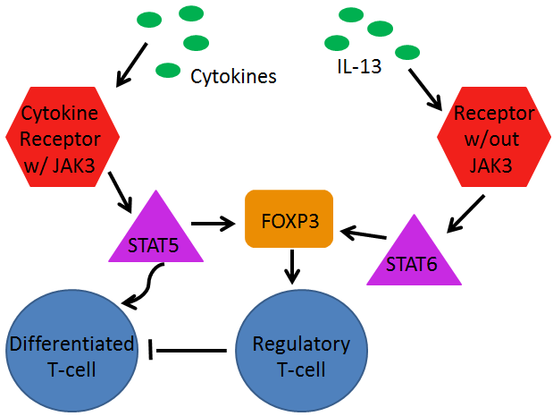
Figure 3 (at left) shows how JAK3 and FOXP3 relate to one another. Just as the diagram HERE shows, cytokines (pictured in green) located outside the cell interact with cytokine receptors (red) located in the cell membranes of progenitor T-cells. Those cytokine receptors may or may not require the presence of JAK3. On the left in the diagram the cytokine receptor uses JAK3 to phosphorylate STAT5 proteins (pink), which can travel into the nucleus of the cell and up-regulate transcription, which will eventually cause the differentiation of the T-cell.
As pictured, the STAT5 protein also plays a part in stabilizing the FOXP3 protein (orange). The FOXP3 protein is activated though through the action of STAT6, which is part of a pathway which does not utilize JAK3. When present FOXP3 will allow regulatory T-cells to develop (blue), which will go on to maintain T-cell homeostasis in the body. [6][7]
If we reduce FOXP3 when JAK3 is deficient, would we be able to maintain a larger number of differentiated T-cells?
Step 1 of the Experiment: Create a JAK3 deficient mouse model
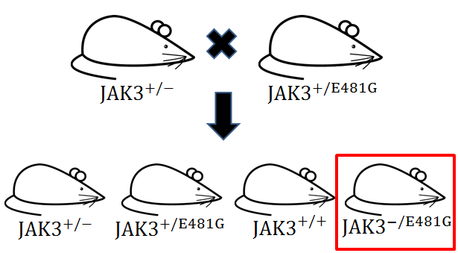
Notarangelo et al. described a child who had one null JAK3 allele and one allele with a mutation labeled E481G. This mutation caused a nucleic acid transition from adenine to guanine which in the protein caused a replacement of a glutamic acid with a glycine. This allele maintained a deficient level of poorly functioning JAK3 in his system. By producing a mouse with this same genotype we would have a model organism expressing an intermediate level of JAK3, being neither wildtype nor completely devoid of JAK3. [8]
To do this we would need to take a heterozygote JAK3 +/- mouse and mate it to a mouse that was heterozygous for the E481G mutation. One out of four resulting mice progeny should present with SCID symptoms. See figure 4 (at right) for a diagram.
To do this we would need to take a heterozygote JAK3 +/- mouse and mate it to a mouse that was heterozygous for the E481G mutation. One out of four resulting mice progeny should present with SCID symptoms. See figure 4 (at right) for a diagram.
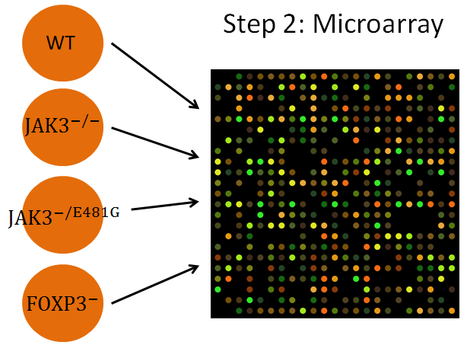
There was no microarray data available that appeared useful for testing this hypothesis at the time this experiment was developed, so step two of testing this hypothesis would involve performing a microarray to correct this. Microarrays would be done using wildtype JAK3 mice (JAK +/+), completely JAK3 deficient mice (JAK3 -/-), mice with an intermediate level of JAK3 (JAK3 -/E481G), and using mice lacking FOXP3 for comparison, looking specifically at the expression of FOXP3 between genotypes. This microarray could also be useful for detecting changes in other proteins that have not previously been identified.
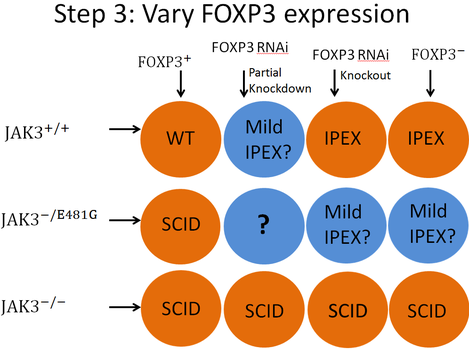
The last experiment which would be useful in testing this hypothesis would involve using the three aforementioned JAK3 varieties of mice and varying the expression of FOXP3 in those mice. A grid of the twelve different strains of mice that would need to be used is located in figure 6 at left. A western blot would need to be performed for each genotype to confirm FOXP3 expression levels. A quantitative T-cell assay would determine T-cell number. Each circle contains an expected result. The blue circles represent results which are not certain. Read below for a description of the "why" behind each expected result. These results are listed in an order that seemed most logical, not in an order that represents the way they are presented in figure 6.
Column 1 Row 1: This would be wild type (WT) genotype as the mice would have normal expression of both proteins.
Column 1 Row 2: This result is based on the assuming the boy from the literature [8] with the E481G had a wild type FOXP3 allele. That boy had SCID, so it is expected these mice should also.
Row 3: All four of the groups of mice represented in row 3 should present with SCID due to their complete lack of JAK3 protein.
Columns 3 & 4, Row 1: Assuming the RNAi knockout works in the mice of column 3, both of these groups of mice should have a complete lack of FOXP3. both groups would be wild type for JAK3, so both groups should present with IPEX.
Columns 3 & 4, Row 2: Again, assuming the RNAi knockout worked for the mice of column 3, both of these groups of mice should have no FOXP3 and should also have an intermediate level of JAK3. No literature was found to propose what phenotype these mice should present with, but I expect they would have either IPEX or a mild form of IPEX.
Column 2 Row 1: There was no literature to suggest a phenotype for this genotype, but with a reduced expression of the FOXP3 protein, but a normal expression of JAK3, it is expected that these mice would have IPEX or a mild form of IPEX.
Column 2 Row 2: This group of mice would potentially be the most important group for testing the hypothesis. In this group both JAK3 and FOXP3 would be deficient. There is no good way to tell at this point if that group of mice would display a wild type phenotype, full SCID, a mild immune deficiency, full IPEX, mild autoimmunity, or some completely new phenotype.
References
[1] Figure 1: http://string.embl.de/newstring_cgi/show_network_section.pl?taskId=4Sput4Tis4q0&advanced_menu=yes&interactive=yes
[2] http://www.ncbi.nlm.nih.gov/gene/50943
[3] Skaguchi, S. (2005). Naturally arising FOXP3-expressing CD25+ CD4+ regulatory T cells in immunological tolerance to self and non-self. Nature Immunology, 6:345-352. doi:10.1038/ni1178
[4] http://www.ncbi.nlm.nih.gov/books/NBK1118/
[5] Figure 2: http://smart.embl-heidelberg.de/smart/set_mode.cgi?GENOMIC=1
[6] Sanchez-Guajardo, V., Tanchot, C., O'Malley, J.T., Kaplan, M.H., Garcia, S., Freitas, A.A.. (2007). Agonist-Driven Development of CD4+ CD25+ FOXP3+ Regulatory T Cells Requires a Second Signal Mediated by Stat6. Journal of Immunology, 178(12):7550-7556. Retrieved from: http://www.jimmunol.org.ezproxy.library.wisc.edu/content/178/12/7550.long
[7] Jang, E., Cho, W.S., Cho, M., Park, H., Oh, H., Kang, S.M., Paik, D., Youn, J. (2011). Foxp3+ Regulatory T Cells Control Humoral Autoimmunity by Suppressing the Development of Long-Lived Plasma Cells. Journal of Immunology, 186(3):1546-1553. doi: 10.4049/jimmunol.1002942
[8] Notarangelo, L.D., Mella, P., Jones, A., de Saint Basile, G., Savoldi, G., Cranston, T., Vihinen, M., Schumacher, R.F. (2001). Mutations in severe combined immune deficiency (SCID) due to JAK3 deficiency. Human Mutation, 18(4):255-63. doi: 10.1002/humu.1188
[9] Figure 3: http://upload.wikimedia.org/wikipedia/commons/c/c9/Simple_mouse.svg
[10] Figure 4: http://commons.wikimedia.org/wiki/File:DNA_microarray.svg
[1] Figure 1: http://string.embl.de/newstring_cgi/show_network_section.pl?taskId=4Sput4Tis4q0&advanced_menu=yes&interactive=yes
[2] http://www.ncbi.nlm.nih.gov/gene/50943
[3] Skaguchi, S. (2005). Naturally arising FOXP3-expressing CD25+ CD4+ regulatory T cells in immunological tolerance to self and non-self. Nature Immunology, 6:345-352. doi:10.1038/ni1178
[4] http://www.ncbi.nlm.nih.gov/books/NBK1118/
[5] Figure 2: http://smart.embl-heidelberg.de/smart/set_mode.cgi?GENOMIC=1
[6] Sanchez-Guajardo, V., Tanchot, C., O'Malley, J.T., Kaplan, M.H., Garcia, S., Freitas, A.A.. (2007). Agonist-Driven Development of CD4+ CD25+ FOXP3+ Regulatory T Cells Requires a Second Signal Mediated by Stat6. Journal of Immunology, 178(12):7550-7556. Retrieved from: http://www.jimmunol.org.ezproxy.library.wisc.edu/content/178/12/7550.long
[7] Jang, E., Cho, W.S., Cho, M., Park, H., Oh, H., Kang, S.M., Paik, D., Youn, J. (2011). Foxp3+ Regulatory T Cells Control Humoral Autoimmunity by Suppressing the Development of Long-Lived Plasma Cells. Journal of Immunology, 186(3):1546-1553. doi: 10.4049/jimmunol.1002942
[8] Notarangelo, L.D., Mella, P., Jones, A., de Saint Basile, G., Savoldi, G., Cranston, T., Vihinen, M., Schumacher, R.F. (2001). Mutations in severe combined immune deficiency (SCID) due to JAK3 deficiency. Human Mutation, 18(4):255-63. doi: 10.1002/humu.1188
[9] Figure 3: http://upload.wikimedia.org/wikipedia/commons/c/c9/Simple_mouse.svg
[10] Figure 4: http://commons.wikimedia.org/wiki/File:DNA_microarray.svg